|
Beckman Research Institute of the City of Hope 1450 E. Duarte Road, Duarte, CA 91010-0269 As far as the English-speaking world is concerned, it seems as though history began with a palindrome. The first self introduction of Adam to Eve in the Garden of Eden was likely to have been "Madam I'm Adam.", which is a perfect palindrome. It should also be noted that EVE, itself, is a palindrome. The beauty of palindromes is found in their perfect symmetry, and because of this symmetry, peptide palindromes appear to have contributed to the functional refinements of diverse proteins (1). In any human language, a perfect and sensible palindrome made of more than 10 words is extremely difficult to compose. The longest that I have come up with, thus far, is: "Wolf's deeds lived was dog DNA, and God saw devil's deeds flow." Two prior knowledge are required for understanding the meaning of the above palindrome. The ancestor of domesticated dogs were wolves and in folklore of the Dark Ages, dogs were often depicted as messengers of the devil. In proteins, on the other hand, peptide palindromes longer than decapeptidic in length are not at all rare. A. 30 Peptidic Palindromes in the 2I2-ResidueLong Mouse HI Histone Variety As shown in Fig. 1, HI histone and other DNA binding proteins tend to be very rich in peptide palindromes. It would be seen that this particular variety known as mouse HI histone variety-1 (2) contained 31 often overlapping peptide palindromes which involved 115 of the 212 residues (54%). The longest palindrome occupied the positions 190-to-203, its tetradecapeptidic palindrome being Lys-Ala-Val-Lys-Pro-Lys-Ala-Ala-Lys-Pro-Lys-Val-Ala-Lys (2nd row from the bottom of Fig. 1). These 31 peptide palindromes, however are not of 31 varieties, for a number of palindromes are found in two or more copies. For example, Lys-Lys-Ala-Ala-Lys-Lys hexapeptidic palindrome occurred twice in the 2nd and 9th row of Fig. 1, while two tripeptidic palindromes made four appearances each; more often than not as parts of longer palindromes. They were Lys-Ala-Lys and Lys-Pro-Lys. The rule of protein construction is that those with unusual amino acid compositions are invariably comprised of oligopeptidic repeats; repeating units often being palindromic (3). In the average amino acid composition deduced from 18,383 entries in DATABASE, the four dominant residues, leu, Ala, Gly and Ser, in the above order, comprise 32% of the total (4). In the case of HI histones, on the contrary, two residues, lys and Ala, in the above order, comprise 48% of the total. With so much deviation from the average, a protein has no choice but to become oligopeptidic repeats. Analogous situations can be found in human languages. Were one asked to compose an essay using a rarely used syllable repeatedly, an essay automatically becomes verse instead of prose. Let me illustrate this point, with a syllable "ze" which is surely one of the seldom used syllables in English. "Zero in Zen cannot be characterized as a void .Rather Zenī s zero is akin t? the absolute zero in temperature, thus representing the symbolized zenith."
Soon after the unveiling of the universal codon assignment, it
became clear that 32 pairs of complementary codons are designed
to encode amino acids of contrasting properties. For example, the
DNA codon CTC (CUC in RNA) encodes leu which is a hydrophobic residue
of substantial size, whereas itS complementary codon GAG encodes
Glu; an acidic therefore, hydrophilic residue. Similarly, the codon
AAG complementary to another Leu codon, CTT, encodes Lys which this
time is a basic but still hydrophilic residue. In fact, contrasts
between amino acids encoded by complementary codons are so striking,
independent suggestions have been made on several occasions, that
each pair of a Peptide hormone and itS receptor must have originally
been encoded by two complementary strands of the same DNA. The disappointing
fact is that complementary strands of coding sequences are, as a
rule, untranslatable in the corresponding reading frame, due to
frequent interruptions by three chain terminators, TAG, TAA and
TGA. Proteins with unusual amino acid compositions are often poor
in leu and Ser and for this reason, complementary strands of coding
sequences encoding these proteins become frequent exceptions to
this rule in that they can be translated in the corresponding reading
frame,to yield proteins that are as long as proteins encoded by
coding sequences. This is because three chain terminators in a complementary
strand are represented in the coding strand as two leu codons, CTA,
TTA and one Ser codon TCA Observing Fig. 1, it would be noted that
the 'coding sequence of mouse HI histone variety-1 is entirely free
of the above three leu and Ser codons. Accordingly, its complementary
strand when translated in .the ,corresponding reading frame, is
capable ?f yielding a protein longer than HI histone, itself. In
Fig. 1, a complementary strand corresponding to HI histone part
are aligned 5'to-3' below the coding sequence and translated in
the corresponding reading frame. Whereas, HI histone was Lys, Ala-rich,
its complementary protein was dominated by Leu, Gly and Arg; the
above three comprising 61% of the total. Yet, this complementary
protein was as rich in peptide palindromes as HI histone itself.
Fig. 1 shows that this complementary protein contained 36 often
overlapping palindromes which involved 115 of the 212 resi,dues
(54%) .Thus, in spite of their contrasting properties, HI histone
and its complementary protein were identical in the portion devoted,
to peptidic palindromes. As to the lengths of palindromes, two octapeptidic
ones, Leu-Arg-Gly-Leu-Leu-Gly-Arg-Leu in the 4th row and Gly Arg-Arg-Leu-Leu-Arg-Arg-Gly
in the 12th row of Fig. 1 were the longest in this complementary
protein. 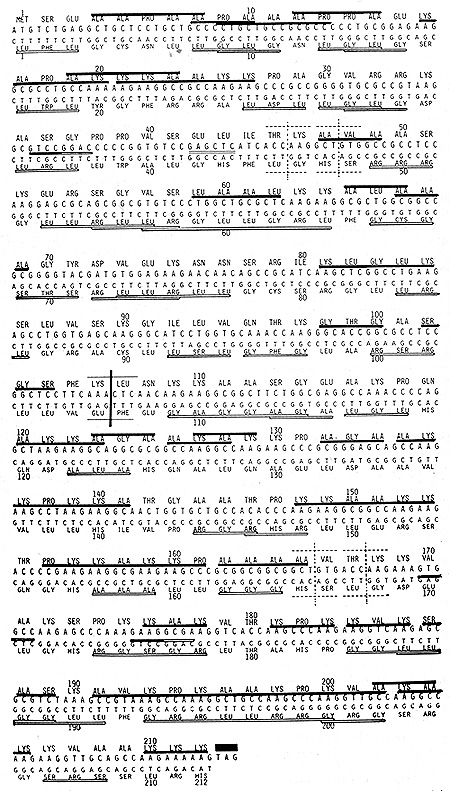
This protein, too, was oligopeptidic repeats. There were four copies of Leu-Gly-Leu tripeptidic palindrome; three becoming a part of Leu-Gly-Leu-Gly repeating tetrapeptides. Another tripeptidic palindrome, Leu-Arg-Leu was also in four copies; two of them becoming a part of the repeating pentapeptidic palindrome, Leu-Leu-Arg-Leu-Leu. Thus, we have learned a few things about a pair of proteins encoded by complementary strands of the same DNA. Ion spite of markedly contrasting properties evidently reflected in their very different amino acid compositions, two proteins share the two basic features: If one is rich in peptide palindromes, so would the other be to an identical degree. If one is made mostly of oligopeptidic repeats, the other would also be of oligopeptidic repeats.
The symmetrical beauty of palindromes was well appreciated by great
composers of the past, notably by Mozart and Haydn. The simultaneous
musical transformation of mouse HI histone variety-l coding sequence
and its open reading frame complementary sequence as the treble
and base clef scores should enable us to appreciate not only the
symmetrical beauty of successive peptide palindromes in HI histone
but also its interplay with equally numerous palindromes in its
complementary protein. This has been done in accordance with the
previously set rule (5). An excerpt of it is shown in Figure 2 that
represented positions I-to-3I followed by positions I48-to-161.
It can be played either on a piano or as a duet between violin and
viola. 
Peptide palindromes are invariably found in all proteins, and
long palindromes exceeding 10 residues in length are not rare. They
are particularly abundant in DNA-binding proteins such as HI histone.
When a complementary strand of the coding sequence is translatable
being free of a chain terminator, a complementary protein encode
by it becomes equally abundant in peptide palindromes. The simultaneous
musical transformation of both strands of mouse H1 histone variety-1
DNA enable us to appreciate the symmetrical beauty of successive
palindromes appearing in both H1 histone and its complementary protein. |