|
1 Institute of Experimental Pathology and Therapy,
USSR Academy of Medical Sciences, Sukhumi, USSR A. Introduction Our previous studies have shown that hamadryas baboons of the Sukhumi
"high lymphoma " stock are infected with human T lymphotrophic virus
(HTL V)-I-related virus to a significantly higher degree than baboons
of different lymphoma-free populations. Levels of anti-HTL V -I-related
antibodies in prelymphomatous baboon sera were also significantly
higher than those in matched controls [1,2]. These studies posed
the question ofwhether HTL V -I-like virus is etiologically related
to baboon malignant lymphoma. The most informative indirect approach
to the study of this possibility is the search for integrated provirus
in baboon lymphoma DNA. Thus, we tested by Southern blotting Pstl,
BamHI, and EcoRI digests of high molecular weight baboon lymphoma
DNA, using as a probe genome-length HTLV-I cloned in SstI site ofpSP-65
vector (this molecular clone was kindly provided by Dr. R. Gallo).
Ten PstI-digested lymphoma DNA samples (lymphomatous lymph nodes)
were found positive (Table 1). The band pattern was similar to,
but clearly different from, that characteristic f or HTL V -I (cf.
Figs. 1, 2, 4). At least three fragments (1.7 kb, 1.5 kb, and 1.1
kb) were observed in all samples (Table 1; Fig. 1 ). They were thought
to be internal fragments [3]. In each positive sample "individual"
bands were also found that suggest monoclonal ( or oligoclonal)
integration of HTL V -I provirus into different sites in baboon
lymphoma DNA (Fig.1). This suggestion was proved correct by Southern
analysis of BamHI and 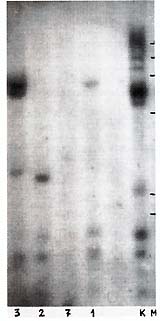
Table I. HTL V -l-related sequences
in DNA of lymphomatous lymph nodes of hamadryas baboons 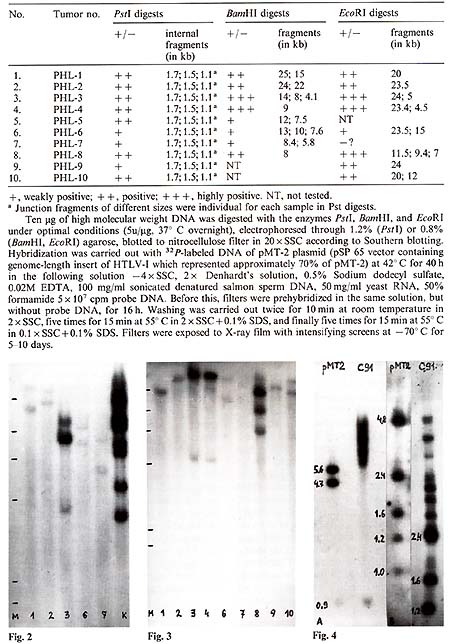
EcoRI digests of the same lymphoma DNA (Table 1; Fig.2, Fig.3). One to three large fragments were observed in each sample, all with individual size. In some cases, fragments were found which were smaller than the expected HTL V -I-like provirus size. These data suggested multiple integration of HTL V -like provirus( es ) in some baboon lymphomas or the oligoclonal origin of these tumors, as well as integration of defective provirus(es) in several cases. HTLV-I-related sequences were not found in one sample of baboon normal lymph node DNA and three samples of muscle DNA isolated from lymphomatous animals.
1. HTLV-I-like provirus is integrated into DNA of baboon malignant
lymphomas.
1. Lapin BA, Voevodill AF, lndzhiia LV et al. (1983) Bull Exp
BioI Med v. XCV, 14-16 (in Russ) |